Our Lab is dedicated to defining the molecular and metabolic mechanisms governing surface invasion by non-motile pathobionts. Currently, we are studying Enterococcus faecalis, a human commensal of the gastrointestinal tract that can cause life-threatening infections. This bacterium has a remarkable ability to overgrow and form dense populations, which can breach intestinal barriers in hosts with disrupted gut homeostasis. This critical process, known as translocation, allows E. faecalis to escape the intestine, invade the bloodstream, and colonize distal organs, drastically altering their physiology.
We use advanced in vitro and in vivo model systems, combined with cutting-edge molecular, biochemical, and genetic approaches, to explore the following exciting questions:
Current projects
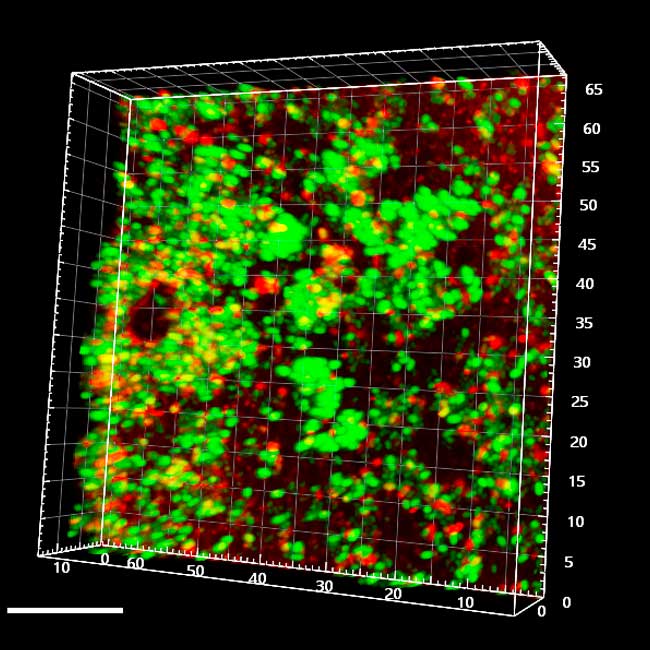
GFP-labelled enterococci covered by red-stained polyGlcNAc
What mechanisms drive bacterial enterococcal aggregate formation during surface penetration?
Our lab found that E. faecalis forms 3D aggregates connected by a self-produced extracellular matrix, partly composed of polyGlcNAc exopolymers, during surface penetration and translocation through intestinal cell barriers. However, the metabolic and physiological states of enterococci within these structures remain poorly understood, and the mechanisms driving their formation are unclear. We determined that E. faecalis cells within these aggregates genetically reprogram their metabolism to enhance cell envelope and glycolipid biogenesis, increasing tolerance to membrane-damaging agents. We are exploring whether surface penetration involves coordinated cell differentiation that influences the spatial structure and physiology of the aggregates. Additionally, we aim to determine if other transcriptional and metabolic programs evolve diverse E. faecalis subpopulations with specialized roles in aggregate development, maintenance, fitness, and penetration.
What metabolic and genetic processes modulate or promote enterococci to penetrate surfaces?
Our work has revealed that Gram-positive cocci, such as Staphylococcus aureus, Streptococcus sp., and E. faecalis, can penetrate and invade diverse surfaces despite not being motile. We found that E. faecalis uses the hexosamine biosynthetic pathway to produce exopolymers mainly composed of poly-N-acetylglucosamine (polyGlcNAc), which are necessary for penetrating both semisolid surfaces and intestinal epithelial cell monolayers. We continue to study the molecular pathways and environmental signals regulating the synthesis of polyGlcNAc and other enterococcal polymers during bacterial movement through surfaces. Moreover, our team is elucidating the cues that trigger penetration/translocation and how these signals are sensed by E. faecalis and other surface-penetrating bacteria
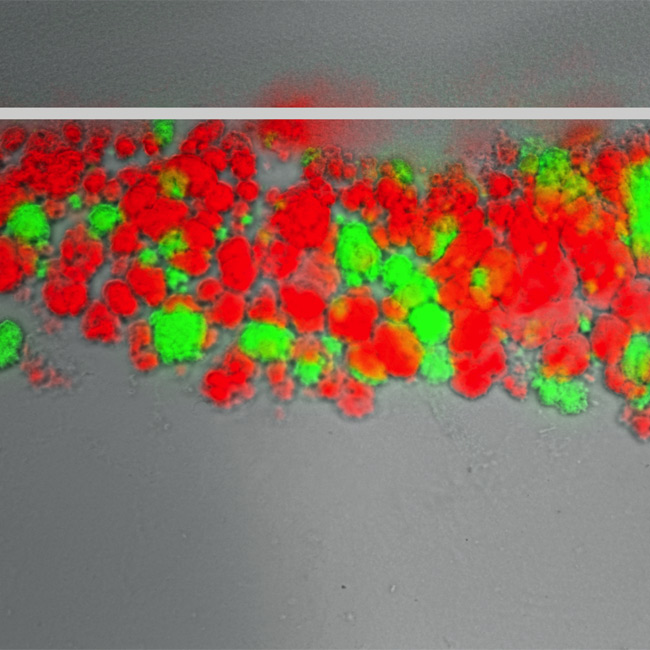
GFP or mCherry-expressing agar-penetrating aggregates

Enterococci aggregating on an intestinal epithelial barrier
Which molecular pathways does E. faecalis disrupt in host cells to alter intestinal barrier integrity and facilitate its translocation?
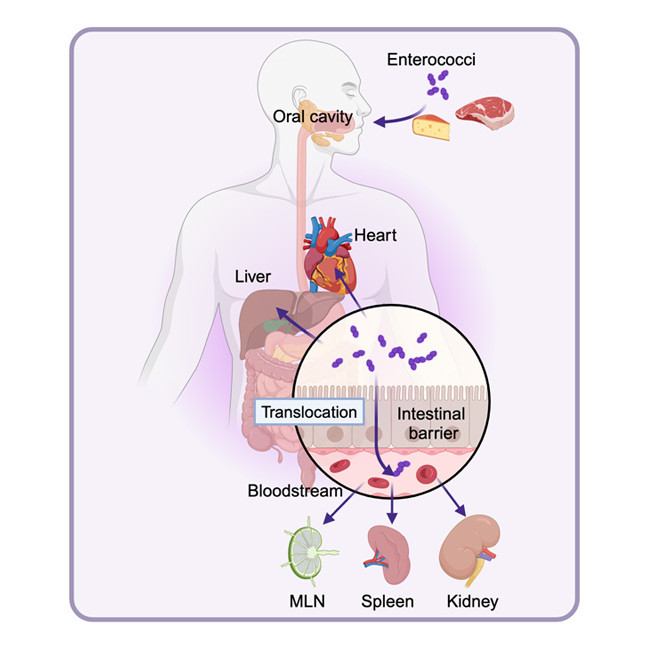
While enterococci mainly inhabit the intestine, factors like diet, chemotherapy, and antibiotics can disrupt gut homeostasis, leading to the overgrowth of multidrug-resistant pathobionts like E. faecalis. Once E. faecalis reaches a critical threshold, it can exit the intestine and spread to other sites. In collaboration with the Cubillos-Ruiz Lab, we are investigating how E. faecalis exploits host responses to breach intestinal barriers. Using murine models, we are exploring the role of the E. faecalis polyGlcNAc-matrix, glycolipid metabolism, and other factors in disrupting intestinal epithelial function and triggering proinflammatory responses. Our focus is on how these non-motile pathobionts manipulate stress pathways in host cells to facilitate their exit from the gut.
How do enterococci interact with host extraintestinal tissues after gut translocation?
When enterococci escape the intestine, they can spread to organs like the liver, spleen, kidneys, and heart, causing infections. Our research focuses on how this microbial translocation disrupts immune cell function in distant tissues. By studying the interactions between E. faecalis and the host immune system, we aim to identify the mechanisms that drive inflammation and physiological changes, worsening infections in these organs. We are also exploring how other gut commensals, especially probiotics, influence the host immune response to E. faecalis, potentially affecting its spread by modulating the local immune environment in the gut.

Human vaginal epithelial cells
How does E. faecalis interact with the female reproductive tract?
Originating from the gastrointestinal tract, E. faecalis can also colonize the reproductive tract of healthy women, with its prevalence rising in those with aerobic vaginitis—a vaginal immune response triggered by the displacement of female commensals by uropathogens, increasing the risk of UTIs. In collaboration with the Department of Urology at Weill Cornell, our lab is investigating the molecular factors that allow enterococci to colonize the female reproductive tract and penetrate vaginal epithelial tissues. We have developed novel models to study E. faecalis colonization of the vaginal tract and the resulting epithelial responses. Our findings show that enterococcal surface penetration induces significant inflammatory responses in host cells, potentially contributing to vaginitis and subsequent UTIs
More about our Research